Volume 8, Issue 2 (Summer 2022)
JMIS 2022, 8(2): 168-183 |
Back to browse issues page
Download citation:
BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:



BibTeX | RIS | EndNote | Medlars | ProCite | Reference Manager | RefWorks
Send citation to:
Ahouz F, Golabpour A, Shakibaeenia A. Proposing a Model for Diagnosing the Type 2 Diabetes Using a Self-Organizing Genetic Algorithm. JMIS 2022; 8 (2) :168-183
URL: http://jmis.hums.ac.ir/article-1-330-en.html
URL: http://jmis.hums.ac.ir/article-1-330-en.html
Department of Health Informatics Technology, School of Allied Medical Sciences, Shahroud University of Medical Sciences, Shahroud, Iran.
Full-Text [PDF 6188 kb]
(804 Downloads)
| Abstract (HTML) (1648 Views)
Full-Text: (943 Views)
Introduction
In recent years, fuzzy systems have been successfully used in different fields of science, especially medical sciences [1, 2]. They use linguistic rules to describe systems that are easily interpreted and checked by the user [3, 4]. Therefore, one of their applications of interest is in clinical decision support systems, where the discovery of rules hidden in the data and the interpretability of these rules are of great importance. The extraction of these rules with two indicators of accuracy and high interpretability helps specialists in increasing the accuracy and speed of disease diagnosis [5, 6, 7].
In the studies that have been conducted so far, attention to the extraction of single rules with high positive predictive value (PPV) and negative predictive value (NPV) has been neglected [8, 9, 10]. Due to the interest of specialists in diagnostic rules that are capable of quick evaluation and are easy to remember, and because many medical data sets, including the Pima diabetes dataset, contain clinical parameters resulting from tests, it is important to design a model for extracting single rules with high efficiency in terms of PPV and NPV. In this paper, we aim to propose a hybrid genetic-fuzzy classification system that automatically extracts the rules hidden in the data. Then, by evaluating each of the extracted rules, we provide the best single rule for diagnosis of disease and absence of disease. In addition, a new self-organizing chromosomal structure is proposed to eliminate the effect of the selection of genetic algorithm operators on the efficiency of the model. The Pima diabetes dataset was finally used to evaluate the proposed model.
Methods
In this study, the rule base of retrospective Mamdani fuzzy systems is designed using PIMA data. One of the problems of PIMA dataset is the presence of missing values and outliers. In this study, KNNi method was used to remove missing values and K-means was applied to remove outliers [2, 9]. At the beginning, there are no rules in the rule base and no membership functions are assigned to the fuzzy variables. They are generated and optimized using the genetic algorithm [2]. To eliminate the effect of the type of mutation and recombination operators on the efficiency of the model and reduce the time of setting the parameters of the genetic algorithm by trial and error, a new chromosomal structure was proposed which, while producing the fuzzy rule base, provides the best mutation and recombination operators among the existing methods for each dataset. In this proposed chromosomal structure, as generations pass and the results converge, the recombination operator corresponding to individuals with the highest fitness is selected more than others. In this way, the optimal recombination operator for the examined data set is automatically selected. The same process is done for the mutation operator. Then, each rule was evaluated on the dataset and their accuracy was measured. Finally, the best single rules with the highest PPV for detecting people with diabetes and the highest NPV for people without diabetes were determined. Afterwards, the diagnostic rules of having and not having the disease were combined and a set of two diagnostic rules was presented as the output of the model.
Results
The result of implementing the proposed model was the presentation of 81 single rules. Among the rules for diagnosing diabetes, those with PPV >70% were selected, and among the rules for diagnosing the absence of diabetes, those with NPV >80% were selected. Four single rules were finally determined which are listed in Table 1.
.jpg)
Table 2 presents the result of combining the best single rules and the output of the proposed model.
.jpg)
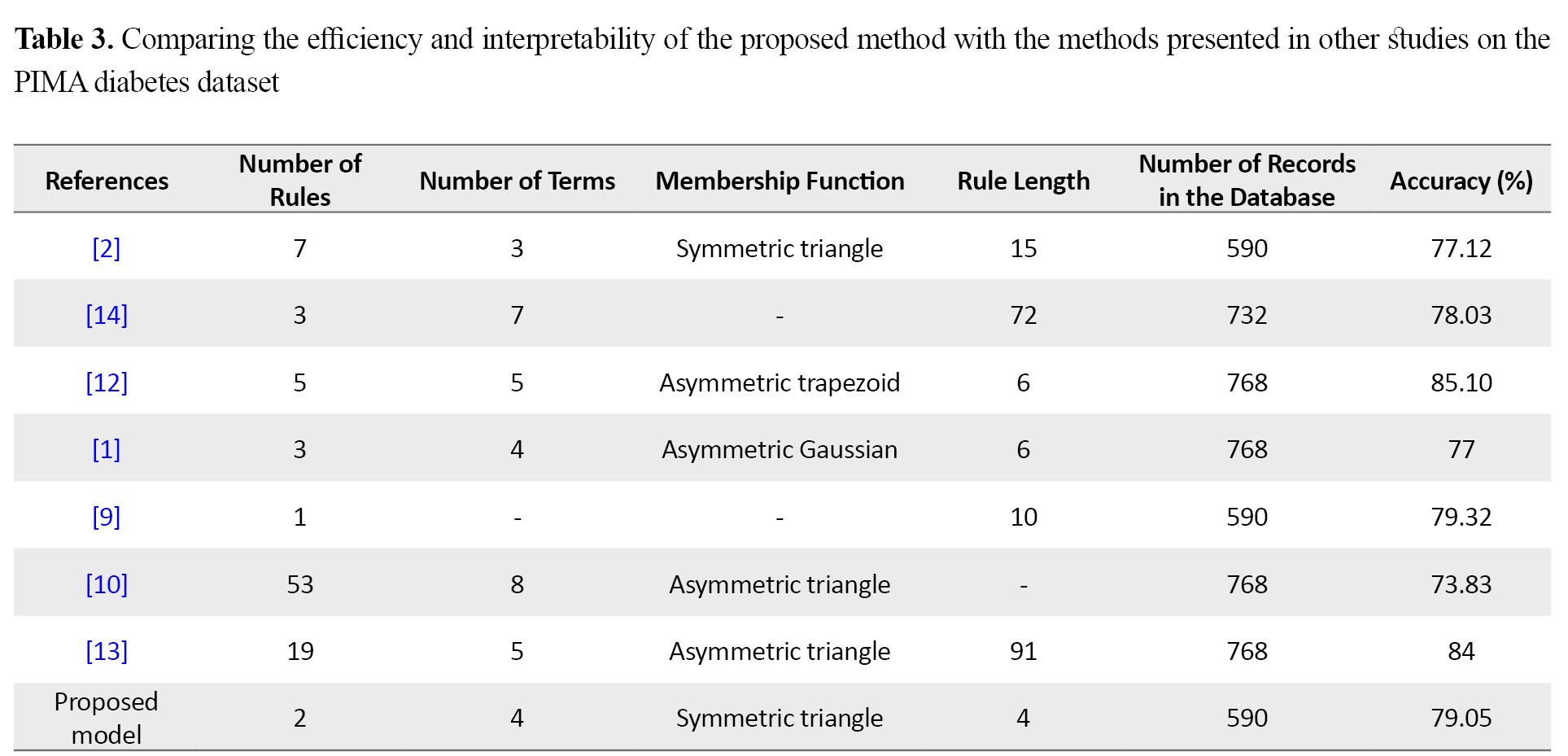
Table 3 compares the efficiency of the proposed method with some existing methods in terms of the number of rules selected in the final rule base, the number of linguistic terms to describe each variable, the fuzzy membership function, the total number of conditions in the rules, the number of records used to build the model, and accuracy.
Discussion
In this study, due to the importance of extracting precise rules and their interpretability in medical assistant systems, a rule extraction model using a hybrid genetic-fuzzy algorithm with high accuracy was presented for determining the most compact set of rules and benchmarked on the PIMA diabetes dataset. In addition, Also, to avoid the complexity of setting genetic algorithm parameters and remove their effect on model efficiency, a new chromosomal structure with automatic adjustment of mutation and recombination operators was presented. The best set of rules according to the two criteria of interpretability and high accuracy, was the set with 2 rules, 4 sets of fuzzy terms per each prefix, and an average length of 2 per each rule, which achieved 79.05% accuracy. The used membership function was a symmetric triangular function, which makes it easier for human users to understand the concepts due to the constant width of all functions.
Based on the rule length criteria, the number of rules and membership functions, which are the most important indicators of the interpretability of rules by human users [10, 11, 12, 13], the proposed model could provide an effective rule set with high interpretability to distinguish diabetic and non-diabetic people including only one diagnostic rule for diabetic people and one diagnostic rule for non-diabetic people.
Conclusion
Rule extraction from datasets in medical diagnosis is an important area for knowledge discovery. Fuzzy systems are known as a popular tool in this application to rules that can be interpreted by humans. For the automatic design of fuzzy systems from the data, the genetic algorithm has shown a high ability. In this study, a self-organizing rule extraction model using genetic-fuzzy system was proposed for diagnostic goals. The PIMA diabetes dataset was used to evaluate the proposed model. Based on this model, the best single diagnostic rules for people without and with diabetes were presented.
Based on the results, it can be concluded that the proposed model with accuracy of 79.05, PPV of 70.83 and NPV of 81.41% can be used as a promising general model in medical data classification and diagnosis. The small number of rules and their shortness are among the important features of the proposed rules, which can be easily evaluated and remembered by experts, along with quick implementation.
One of the limitations of the study was the use of public data and not evaluation of the proposed rules by clinical experts. It is suggested that the proposed model be implemented on local data and each of the extracted rules be evaluated by clinical experts.
Ethical Considerations
Compliance with ethical guidelines
In this study, no experiment on human or animal samples were conducted. Therefore, there as no need for obtaining ethical code.
Funding
This study was extracted from a research project (Code: 400-01-00) in Behbahan Khatam Alanbia University of Technology. It was not funded by any organization.
Authors' contributions
Conceptualization: Amin Golabpour; Writing: Fatemeh Ahouz; Methodology, model implementation and analysis: All authors
Conflicts of interest
The authors declared no conflict of interest.
References
In recent years, fuzzy systems have been successfully used in different fields of science, especially medical sciences [1, 2]. They use linguistic rules to describe systems that are easily interpreted and checked by the user [3, 4]. Therefore, one of their applications of interest is in clinical decision support systems, where the discovery of rules hidden in the data and the interpretability of these rules are of great importance. The extraction of these rules with two indicators of accuracy and high interpretability helps specialists in increasing the accuracy and speed of disease diagnosis [5, 6, 7].
In the studies that have been conducted so far, attention to the extraction of single rules with high positive predictive value (PPV) and negative predictive value (NPV) has been neglected [8, 9, 10]. Due to the interest of specialists in diagnostic rules that are capable of quick evaluation and are easy to remember, and because many medical data sets, including the Pima diabetes dataset, contain clinical parameters resulting from tests, it is important to design a model for extracting single rules with high efficiency in terms of PPV and NPV. In this paper, we aim to propose a hybrid genetic-fuzzy classification system that automatically extracts the rules hidden in the data. Then, by evaluating each of the extracted rules, we provide the best single rule for diagnosis of disease and absence of disease. In addition, a new self-organizing chromosomal structure is proposed to eliminate the effect of the selection of genetic algorithm operators on the efficiency of the model. The Pima diabetes dataset was finally used to evaluate the proposed model.
Methods
In this study, the rule base of retrospective Mamdani fuzzy systems is designed using PIMA data. One of the problems of PIMA dataset is the presence of missing values and outliers. In this study, KNNi method was used to remove missing values and K-means was applied to remove outliers [2, 9]. At the beginning, there are no rules in the rule base and no membership functions are assigned to the fuzzy variables. They are generated and optimized using the genetic algorithm [2]. To eliminate the effect of the type of mutation and recombination operators on the efficiency of the model and reduce the time of setting the parameters of the genetic algorithm by trial and error, a new chromosomal structure was proposed which, while producing the fuzzy rule base, provides the best mutation and recombination operators among the existing methods for each dataset. In this proposed chromosomal structure, as generations pass and the results converge, the recombination operator corresponding to individuals with the highest fitness is selected more than others. In this way, the optimal recombination operator for the examined data set is automatically selected. The same process is done for the mutation operator. Then, each rule was evaluated on the dataset and their accuracy was measured. Finally, the best single rules with the highest PPV for detecting people with diabetes and the highest NPV for people without diabetes were determined. Afterwards, the diagnostic rules of having and not having the disease were combined and a set of two diagnostic rules was presented as the output of the model.
Results
The result of implementing the proposed model was the presentation of 81 single rules. Among the rules for diagnosing diabetes, those with PPV >70% were selected, and among the rules for diagnosing the absence of diabetes, those with NPV >80% were selected. Four single rules were finally determined which are listed in Table 1.
.jpg)
Table 2 presents the result of combining the best single rules and the output of the proposed model.
.jpg)
Table 3 compares the efficiency of the proposed method with some existing methods in terms of the number of rules selected in the final rule base, the number of linguistic terms to describe each variable, the fuzzy membership function, the total number of conditions in the rules, the number of records used to build the model, and accuracy.

Discussion
In this study, due to the importance of extracting precise rules and their interpretability in medical assistant systems, a rule extraction model using a hybrid genetic-fuzzy algorithm with high accuracy was presented for determining the most compact set of rules and benchmarked on the PIMA diabetes dataset. In addition, Also, to avoid the complexity of setting genetic algorithm parameters and remove their effect on model efficiency, a new chromosomal structure with automatic adjustment of mutation and recombination operators was presented. The best set of rules according to the two criteria of interpretability and high accuracy, was the set with 2 rules, 4 sets of fuzzy terms per each prefix, and an average length of 2 per each rule, which achieved 79.05% accuracy. The used membership function was a symmetric triangular function, which makes it easier for human users to understand the concepts due to the constant width of all functions.
Based on the rule length criteria, the number of rules and membership functions, which are the most important indicators of the interpretability of rules by human users [10, 11, 12, 13], the proposed model could provide an effective rule set with high interpretability to distinguish diabetic and non-diabetic people including only one diagnostic rule for diabetic people and one diagnostic rule for non-diabetic people.
Conclusion
Rule extraction from datasets in medical diagnosis is an important area for knowledge discovery. Fuzzy systems are known as a popular tool in this application to rules that can be interpreted by humans. For the automatic design of fuzzy systems from the data, the genetic algorithm has shown a high ability. In this study, a self-organizing rule extraction model using genetic-fuzzy system was proposed for diagnostic goals. The PIMA diabetes dataset was used to evaluate the proposed model. Based on this model, the best single diagnostic rules for people without and with diabetes were presented.
Based on the results, it can be concluded that the proposed model with accuracy of 79.05, PPV of 70.83 and NPV of 81.41% can be used as a promising general model in medical data classification and diagnosis. The small number of rules and their shortness are among the important features of the proposed rules, which can be easily evaluated and remembered by experts, along with quick implementation.
One of the limitations of the study was the use of public data and not evaluation of the proposed rules by clinical experts. It is suggested that the proposed model be implemented on local data and each of the extracted rules be evaluated by clinical experts.
Ethical Considerations
Compliance with ethical guidelines
In this study, no experiment on human or animal samples were conducted. Therefore, there as no need for obtaining ethical code.
Funding
This study was extracted from a research project (Code: 400-01-00) in Behbahan Khatam Alanbia University of Technology. It was not funded by any organization.
Authors' contributions
Conceptualization: Amin Golabpour; Writing: Fatemeh Ahouz; Methodology, model implementation and analysis: All authors
Conflicts of interest
The authors declared no conflict of interest.
References
- Ishibuchi H, Nojima Y, Kuwajima I. Genetic rule selection as a postprocessing procedure in fuzzy data mining. Int Symp Evolv Fuzzy Syst; 2006. [DOI:10.1109/ISEFS.2006.251149]
- Ahouz F, Golabpour A. A novel compact rule extractor based on genetic-fuzzy algorithm. Paper presented at: 10th International Conference on Computer and Knowledge Engineering (ICCKE); 29-30 October 2020, Mashhad, Iran. [DOI:10.1109/ICCKE50421.2020.9303613]
- Gorzałczany MB, Rudziński F. Interpretable and accurate medical data classification–a multi-objective genetic-fuzzy optimization approach. Expert Syst Appl. 2017; 71:26-39. [DOI:10.1016/j.eswa.2016.11.017]
- Mitra S, Hayashi Y. Neuro-fuzzy rule generation: Survey in soft computing framework. IEEE Trans Neural Netw. 2000; 11(3):748-68. [PMID]
- Shi Y, Eberhart R, Chen Y. Implementation of evolutionary fuzzy systems. IEEE Trans Fuzzy Syst. 1999;7(2):109-19. [DOI:10.1109/91.755393]
- Ahouz F, Golabpour A. A novel structure of highly interpretable fuzzy rules extraction. Front Health Inform. 2021; 10(1):53. [DOI:10.30699/fhi.v10i1.253]
- Shortliffe EH, Wiederhold G, Fagan LM, Perreault LE. Medical Informatics: Computer Applications in Health Care and Biomedicine. New York: Springer; 2013. [Link]
- Sujatha R, Ephzibah EP, Dharinya S, Uma Maheswari G, Mareeswari V, Pamidimarri V. Comparative study on dimensionality reduction for disease diagnosis using fuzzy classifier. Int J Eng Technol. 2018; 7(1):79-84.[DOI:10.14419/ijet.v7i1.8652]
- Seera M, Lim CP. A hybrid intelligent system for medical data classification. Expert Syst Appl. 2014; 41(5):2239-49. [DOI:10.1016/j.eswa.2013.09.022]
- Chang X, Lilly JH. Evolutionary design of a fuzzy classifier from data. IEEE Trans Syst Man Cybern B Cybern. 2004; 34(4):1894-906. [PMID]
- GaneshKumar P, Rani C, Devaraj D, Victoire TAA. Hybrid ant bee algorithm for fuzzy expert system based sample classification. IEEE ACM Trans Comput Biol Bioinformatics. 2014; 11(2):347-60. [PMID]
- Pinto CMA, Carvalho ARM. Diabetes mellitus and TB co-existence: Clinical implications from a fractional order modelling. Appl Math Model. 2019; 68:219-43. [DOI:10.1016/j.apm.2018.11.029]
- Abedini S, Jomehpour S, Fallahi S, Ghanbarnejad A, Nikparvar M. [The effect of virtual education of cardiovascular risk factors on the knowledge of general physicians (Persian)]. J Mod Med Inf Sci. 2020; 6(1):31-6. [DOI:10.29252/jmis.6.1.32]
- Schulz LO, Chaudhari LS. High-risk populations: The pimas of Arizona and Mexico. Curr Obes Rep. 2015; 4(1):92-8. [PMID]
- Ahouz F, Sadehvand M, Golabpour A. Extracting rules for diagnosis of diabetes using genetic programming. Int J Health Stud. 2019; 5(3):23-32. [DOI:10.22100/ijhs.v5i3.691]
- Feng TC, Li THS, Kuo PH. Variable coded hierarchical fuzzy classification model using DNA coding and evolutionary programming. Appl Math Model. 2015; 39(23-24):7401-19. [DOI:10.1016/j.apm.2015.03.004]
- Singh S, Singh S, Banga VK. Design of fuzzy logic system framework using evolutionary techniques. Soft Comput. 2020; 24(6):4455-68. [DOI:10.1007/s00500-019-04207-9]
- López-Campos JA, Segade A, Fernández JR, Casarejos E, Vilán J. Behavior characterization of visco-hyperelastic models for rubber-like materials using genetic algorithms. Appl Math Model. 2019; 66:241-55. [DOI:10.1016/j.apm.2018.08.031]
- Mansourypoor F, Asadi S. Development of a reinforcement learning-based evolutionary fuzzy rule-based system for diabetes diagnosis. Comput Biol Med. 2017; 91:337-52. [PMID]
- Tan CH, Tan MS, Chang SW, Yap KS, Yap HJ, Wong SY. Genetic algorithm fuzzy logic for medical knowledge-based pattern classification. J Eng Sci Technol. 2018; 13:242-58. [Link]
- Vaishali R, Sasikala R, Ramasubbareddy S, Remya S, Nalluri S. Genetic algorithm based feature selection and MOE Fuzzy classification algorithm on Pima Indians Diabetes dataset. International Conference on Computing Networking and Informatics (ICCNI); 29-30 October 2017; Lagos, Nigeria. [DOI:10.1109/ICCNI.2017.8123815]
Type of Study: Research |
Subject:
Special
Received: 2021/07/30 | Accepted: 2022/05/8 | Published: 2022/07/1
Received: 2021/07/30 | Accepted: 2022/05/8 | Published: 2022/07/1
Send email to the article author
| Rights and permissions | |
 |
This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License. |








 hums.ac.ir
hums.ac.ir